Wpływ zanieczyszczenia gleby na występowanie i różnorodność przedstawicieli Mucoromycota oraz ich bakteryjnych endosymbiontów
 Pleśniakowce (Mucoromycota) są grzybami powszechnie spotykanymi w różnych siedliskach na całym świecie. Można je spotkać na pleśniejącym chlebie czy popsutych truskawkach. Znakomita większość z nich to organizmy rozkładające martwą materię organiczną. W ostatnich latach okazało się, że wewnątrz strzępek niektórych pleśniakowców mogą występować bakterie. Ich różnorodność oraz czynniki wpływające na częstość ich występowania pozostają jednak słabo poznane. Celem projektu było zbadanie wpływu zanieczyszczenia gleby na występowanie i różnorodność grzybów oraz na obecność bakterii mogących zasiedlać strzępki przedstawicieli Mucoromycota.
Pleśniakowce (Mucoromycota) są grzybami powszechnie spotykanymi w różnych siedliskach na całym świecie. Można je spotkać na pleśniejącym chlebie czy popsutych truskawkach. Znakomita większość z nich to organizmy rozkładające martwą materię organiczną. W ostatnich latach okazało się, że wewnątrz strzępek niektórych pleśniakowców mogą występować bakterie. Ich różnorodność oraz czynniki wpływające na częstość ich występowania pozostają jednak słabo poznane. Celem projektu było zbadanie wpływu zanieczyszczenia gleby na występowanie i różnorodność grzybów oraz na obecność bakterii mogących zasiedlać strzępki przedstawicieli Mucoromycota.
W ramach realizacji projektu przebadaliśmy glebę z 50 stanowisk z całej Polski. Stanowiska były dobrane w taki sposób, aby połowa z nich reprezentowała obszary silnie zanieczyszczone, np. hałdy pokopalniane. Każda próba gleby została scharakteryzowana pod względem stopnia i typu zanieczyszczenia. W oparciu o metody sekwencjonowania nowej generacji we wszystkich próbach zbadano też różnorodność gatunkową grzybów, a ponadto – na wybranych dziesięciu – również bakterii. Badania te pozwoliły wykazać, że zanieczyszczenie m.in. wielopierścieniowymi węglowodorami aromatycznymi jest czynnikiem kształtującym różnorodność zbiorowisk grzybów glebowych.
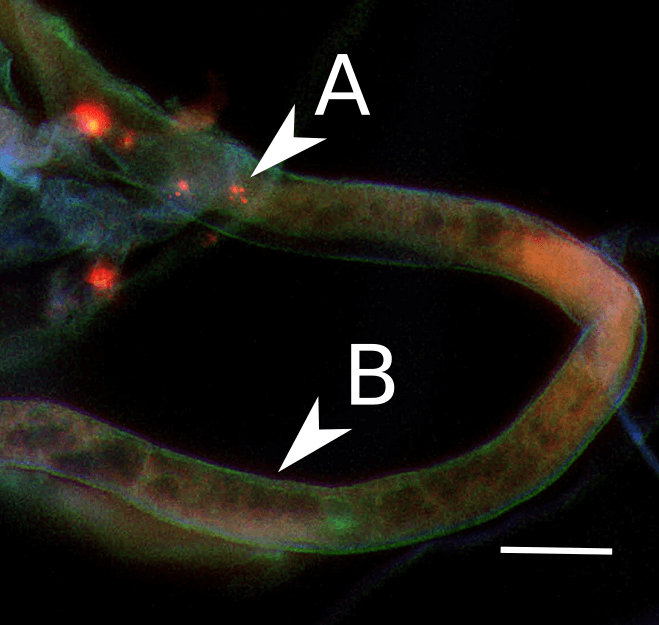
Ponadto, z wybranych 10 prób gleby wyizolowano przedstawicieli pleśniakowców i sprawdzono czy w ich strzępkach występują bakterie. Ich obecność stwierdzono u ok. 20% wszystkich wyizolowanych szczepów grzybów (Rys. 1.). Wśród zidentyfikowanych endosymbiontów odkryto nową linię bakterii wewnątrzstrzępkowych. W ramach realizacji projektu odkryto i opisano też dwa nowe dla nauki gatunki grzybów.
Na koniec, spośród szczepów posiadających bakterie wewnątrzstrzępkowe wybrano dwa, które pozbawiono bakterii, a następnie sprawdzono ich zdolność do wzrostu na wybranych zanieczyszczeniach. Wykazano, że szczepy Umbelopsis oraz Rhizopus zawierające bakterie wewnątrzstrzępkowe są zdolne do degradacji oleju napędowego.
Podsumowując, przeprowadzone w ramach realizacji grantu badania pozwoliły wykazać wpływ zanieczyszczenia gleby na skład gatunkowy grzybów oraz zbadać różnorodność bakterii wewnątrzstrzępkowych u przedstawicieli pleśniakowców. Niektóre ze zidentyfikowanych bakterii wydają się wpływać na zdolność adaptacji grzybowych gospodarzy do specyficznych warunków środowiskowych, takich jak obecność zanieczyszczeń.
Główne wyniki:
- Opisano nowy gatunek Sugiyamaella trypani A. Gęsiorska & J. Pawłowska [MB#829450] ze zlewek zanieczyszczonych błękitem trypanu.
- Opisano nowy gatunek Entomortierella hereditatis J. Trovão, J. Pawłowska & A. Portugal [MB#825064].
- Odkryto nową linię bakterii wewnątrzstrzępkowych u przedstawicieli rzędu Umbelopsidales.
- Wykazano koewolucję pomiędzy bakteriami wewnątrzstrzępkowymi należącymi do BRE a ich grzybowymi gospodarzami należącymi do Mucoromycota.
- Wykazano, że obecność bakterii wewnątrzstrzępkowych może wpływać na zdolności metaboliczne grzybowych gospodarzy.
- Wykazano, że zanieczyszczenie gleby m.in. wielopierścieniowymi węglowodorami aromatycznymi jest czynnikiem kształtującym różnorodność zbiorowisk grzybów.
- Wykazano, że wybrane szczepy Umbelopsis oraz Rhizopus zawierające bakterie wewnątrzstrzępkowe są zdolne do degradacji oleju napędowego oraz wykorzystywania antracenu, undekanu i cykloheksanu jako jedynych źródeł węgla.
Publikacje:
2024
Czy grzyby mają swój mikrobiom? O bakteriach żyjących w grzybowych strzępkach Journal Article
In: Kosmos Problemy Nauk Biologicznych, vol. 73, iss. 3 (2024), pp. 369–378, 2024.
Fungal Planet description sheets: 1614–1696 Journal Article
In: Fungal Systematics and Evolution, vol. 13, no. 11, pp. 183-440, 2024.
Mucor thermorhizoides—A New Species from Post-mining Site in Sudety Mountains (Poland). Journal Article
In: Current Microbiology, vol. 81, iss. 201, 2024.
The interkingdom horizontal gene transfer in 44 early diverging fungi boosted their metabolic, adaptive, and immune capabilities Journal Article
In: Evolution Letters, vol. 03, pp. qrae009, 2024, ISSN: 2056-3744.
2022
Marginal lands and fungi – linking the type of soil contamination with fungal community composition Journal Article
In: Environmental Microbiology, 2022.
2021
Fungal Planet description sheets: 1284–1382 Journal Article
In: Persoonia, vol. 47, pp. 309, 2021, ISSN: 1878-9080.
Metabolic Potential, Ecology and Presence of Associated Bacteria Is Reflected in Genomic Diversity of Mucoromycotina Journal Article
In: Frontiers in Microbiology, vol. 12, pp. 239, 2021.
New Endohyphal Relationships between Mucoromycota and Burkholderiaceae Representatives Journal Article
In: Applied and Environmental Microbiology, vol. 87, no. 7, 2021, ISSN: 0099-2240.
2020
Outline of Fungi and fungus-like taxa Journal Article
In: Mycosphere, vol. 11, no. 1, pp. 1060–1456, 2020, ISSN: 2077-7000.
2019
Carbon assimilation profiles of mucoralean fungi show their metabolic versatility Journal Article
In: Scientific Reports, vol. 9, pp. 11864, 2019.
Fungal Planet description sheets: 868-950 Journal Article
In: Persoonia, vol. 42, pp. 291, 2019.
2018
In: Fungal Diversity, vol. 92, no. 1, pp. 43–129, 2018, ISSN: 1560-2745.
Prezentowanie wyników na konferencjach:
1. J. Pawłowska, A. Okrasińska, P. Decewicz, Ł. Dziewit: Wpływ zanieczyszczenia na różnorodność zbiorowisk grzybów glebowych. Konferencja “Metagenomy różnych środowisk”, on-line, 17-18.06.2021 – prezentacja.
2. Z. Błocka: Analysis of Diesel Oil Degradation by Fungi and Endohyphal Bacteria. CYEEB, 3-5.9.21 – prezentacja.
3. A. Okrasińska: Bakterie wewnątrzstrzępkowe grzybów Mucoromycota. MycoRiseUp! Młodzi w mykologii, on-line, 23-25.04.2021 – prezentacja.
4. A. Gęsiorska: Diversity of endosymbiotic bacteria in Umbelopsis representatives. MycoRiseUp! Młodzi w mykologii, on-line, 23-25.04.2021 – prezentacja.
5. A. Okrasińska, P. Decewicz, J. Pawłowska: Linking soil contamination type with fungal community composition. Trends in Biodiversity and Evolution Conference (https://cibio.up.pt/tibe/details/tibe2020), on-line, 9-11.12.2020 – prezentacja.
6. J. Pawłowska, P. Decewicz, A. Okrasińska, Ł. Dziewit, M. Wrzosek: Soil contamination as a key factor influencing fungal diversity. 18th Congress of European Mycologists, Warszawa-Białowieża (Polska), 16-21.9.2019 – poster.
7. A. Okrasińska, A. Bokus, A. Gęsiorska, B. Sokołowska, M. Wrzosek, J. Pawłowska: Bacterial endosymbionts of Mucoromycota representatives, 18th Congress of European Mycologists, Warszawa-Białowieża (Polska), 16-21.9.2019 – poster.
8. J. Pawłowska, A. Okrasińska, K. Duk, Ł. Istel, M. Gorczak, I. Siedlecki, A. Bokus, A. Gęsiorska, M. Wrzosek: Diversity of endohyphal bacteria in Mucoromycota representatives. 58. Zjazd Polskiego Towarzystwa Botanicznego, Kraków (Polska), 1-7.7.2019 – prezentacja.
9. A.Okrasińska, A. Bokus, A. Gęsiorska, M. Wrzosek, J. Pawłowska: Could have fungal-bacterial interactions helped plants in land colonisation? PopBio, Warszawa (Polska), 22-25.05.2019 – prezentacja.
10. A. Okrasińska, K. Duk, A. Bokus, J. Pawłowska: New lineages of endohyphal bacteria detected in land colonizing fungi. 6th Young Natural History scientists Meeting, Paryż (Francja), 12-16.3.2019 – poster.
11. A. Gęsiorska, J. Pawłowska: Sugiyamaella trypani – new species isolated from trypan blue dye. 6th Young Natural History scientists Meeting, Paryż (Francja), 12-16.3.2019 – poster.
12. J. Pawłowska , A. Okrasińska, L. Serewa, A. Miłobędzka, G. Ostrowski, Ł. Dziewit, P. Decewicz, M. Wrzosek: Influence of soil contamination on the occurence and diversity of Mucoromycota and their endosymbiotic bacteria. Bacterial-Fungal Interactions Workshop during 11 International Mycological Congress, San Juan (Puerto Rico), 16-21.07.2018 – prezentacja.
Wydarzenia popularnonaukowe:
1. film “o magii poszukiwania”: https://www.youtube.com/watch?v=VlWni46_wmI
2. udział w stanowisku WB UW na Pikniku Naukowym Polskiego Radia i Centrum Nauki Kopernik (2020)
3. udział w Nocy Biologów (2020)
4. udział w Dniu Odkrywców Kampusu Ochota (2019)
5. prezentacja na Targach Eurolab (2019)
——————————————————————————————————————————————————————————————
